5 Null models
Jordi Bascompte & Fernando Pedraza
session 24/03/2022
5.1 Null models by hand
In this section you will perform a randomization by hand of the provided networks using the null models covered during our morning lecture. Use R to generate random numbers (using the runif() function) and for your operations.
- Consider a network represented by the following adjacency matrix:
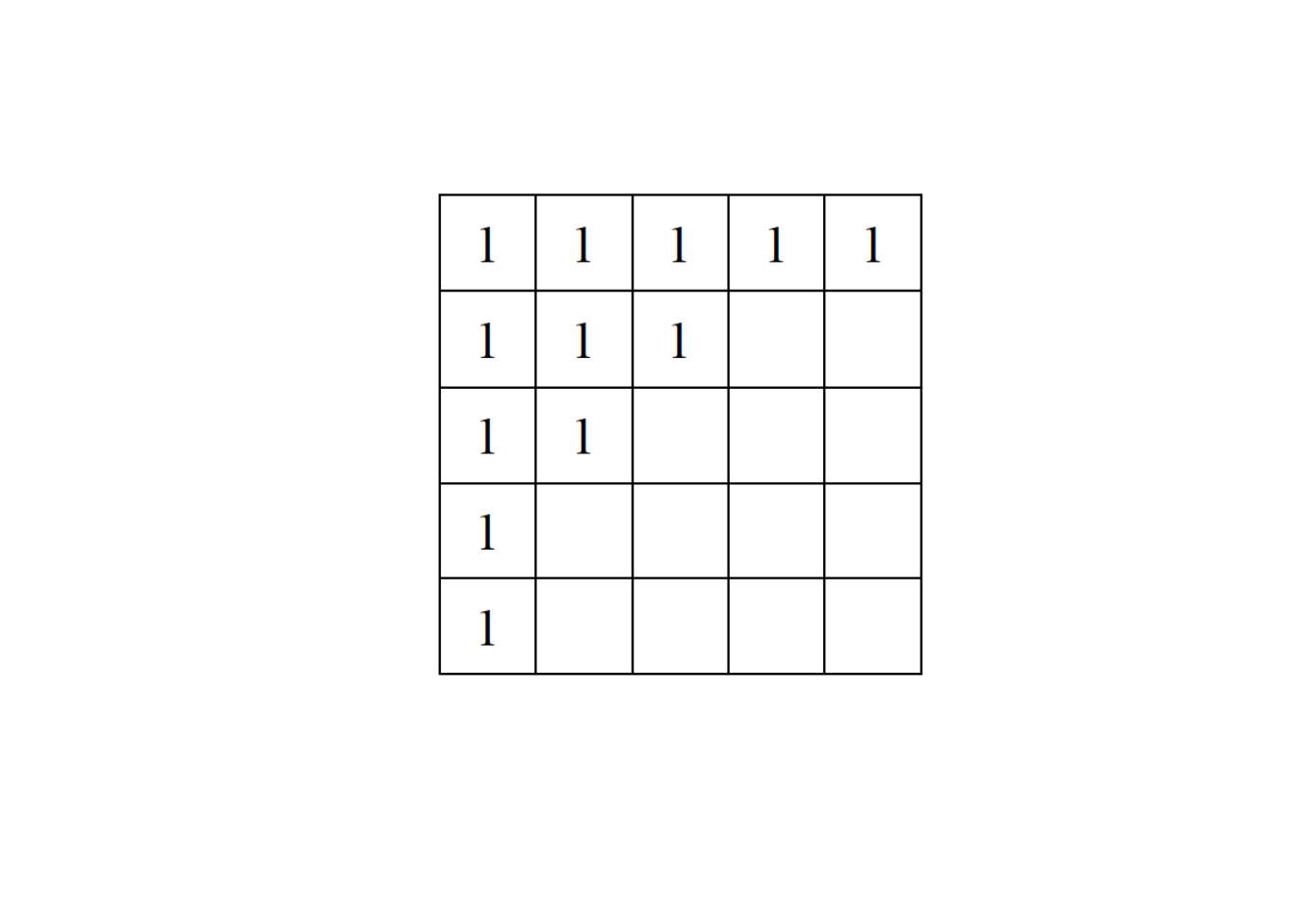
Provide a randomization of the matrix using:
a. the equifrequent null model
In your Rscript file, replace the ’x’s with the numbers you came up with in the randomization for the cell null model.
# -----------------------------------------------------------
#| | | | | |
#| x | x | x | x | x |
#| | | | | |
#|-----------------------------------------------------------|
#| | | | | |
#| x | x | x | x | x |
#| | | | | |
#|-----------------------------------------------------------|
#| | | | | |
#| x | x | x | x | x |
#| | | | | |
#|-----------------------------------------------------------|
#| | | | | |
#| x | x | x | x | x |
#| | | | | |
#|-----------------------------------------------------------|
#| | | | | |
#| x | x | x | x | x |
#| | | | | |
# -----------------------------------------------------------b. the probabilistic cell null model
In your Rscript file, replace the ’x’s with the numbers you came up with in the randomization for the cell null model.
# -----------------------------------------------------------
#| | | | | |
#| x | x | x | x | x |
#| | | | | |
#|-----------------------------------------------------------|
#| | | | | |
#| x | x | x | x | x |
#| | | | | |
#|-----------------------------------------------------------|
#| | | | | |
#| x | x | x | x | x |
#| | | | | |
#|-----------------------------------------------------------|
#| | | | | |
#| x | x | x | x | x |
#| | | | | |
#|-----------------------------------------------------------|
#| | | | | |
#| x | x | x | x | x |
#| | | | | |
# ------------------------------------------------------------ Consider now a second network represented by this adjacency matrix:
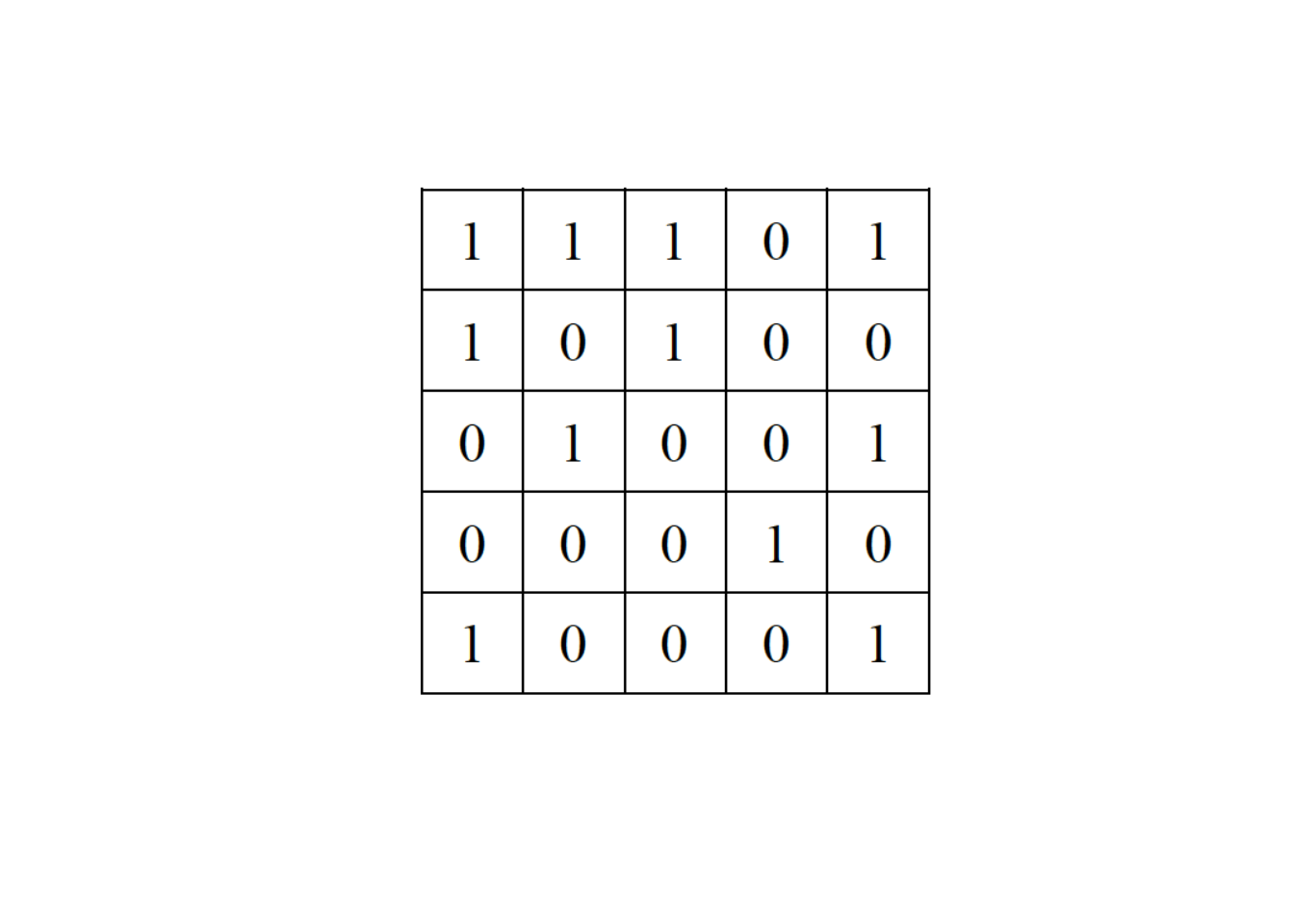
Provide three iterations of the swap algorithm:
first iteration
In your Rscript file, replace the ’x’s with the numbers you came up with in the randomization for the cell null model.
# -----------------------------------------------------------
#| | | | | |
#| x | x | x | x | x |
#| | | | | |
#|-----------------------------------------------------------|
#| | | | | |
#| x | x | x | x | x |
#| | | | | |
#|-----------------------------------------------------------|
#| | | | | |
#| x | x | x | x | x |
#| | | | | |
#|-----------------------------------------------------------|
#| | | | | |
#| x | x | x | x | x |
#| | | | | |
#|-----------------------------------------------------------|
#| | | | | |
#| x | x | x | x | x |
#| | | | | |
# -----------------------------------------------------------second iteration
In your Rscript file, replace the ’x’s with the numbers you came up with in the randomization for the cell null model.
# -----------------------------------------------------------
#| | | | | |
#| x | x | x | x | x |
#| | | | | |
#|-----------------------------------------------------------|
#| | | | | |
#| x | x | x | x | x |
#| | | | | |
#|-----------------------------------------------------------|
#| | | | | |
#| x | x | x | x | x |
#| | | | | |
#|-----------------------------------------------------------|
#| | | | | |
#| x | x | x | x | x |
#| | | | | |
#|-----------------------------------------------------------|
#| | | | | |
#| x | x | x | x | x |
#| | | | | |
# -----------------------------------------------------------third iteration
In your Rscript file, replace the ’x’s with the numbers you came up with in the randomization for the cell null model.
# -----------------------------------------------------------
#| | | | | |
#| x | x | x | x | x |
#| | | | | |
#|-----------------------------------------------------------|
#| | | | | |
#| x | x | x | x | x |
#| | | | | |
#|-----------------------------------------------------------|
#| | | | | |
#| x | x | x | x | x |
#| | | | | |
#|-----------------------------------------------------------|
#| | | | | |
#| x | x | x | x | x |
#| | | | | |
#|-----------------------------------------------------------|
#| | | | | |
#| x | x | x | x | x |
#| | | | | |
# -----------------------------------------------------------5.2 Computer part I
In this section we will use R code to run the null models covered during this morning’s lecture. We will focus on using the null models to evaluate the nestedness value of a single network. Ultimately, we want to know if the measured nestedness value of our network is significantly different from the nestedness values obtained from a set of random iterations. To do this, we will first download a network and compute it’s nestedness. Then, we’ll compute nestedness again using the three null models we went over. Finally, we will estimate the significance of nestedness using the z-score.
Before we begin we should load the packages we will use for this sesssion.
# Load packages
library(igraph)
library(data.table)
library(tidyverse)
library(Matrix)
library(rjson) 5.2.1 Downloading a network
First we will download the network called “M_PL_011” as you have done in the previous lectures.
# define the base url
base_url <- "https://www.web-of-life.es/"
# define the network name
net_name <- 'M_PL_011'
# define the full url associated with the endpoint that download networks interactions
json_url <- paste0(base_url,"get_networks.php?network_name=",net_name)
# and run it
M_PL_011 <- jsonlite::fromJSON(json_url)
# select the 3 relevant column and pass and create the igraph object
M_PL_011_graph <- M_PL_011 %>% select(species1, species2, connection_strength) %>%
graph_from_data_frame(directed = FALSE)
# convert network to bipartite
V(M_PL_011_graph)$type <- bipartite_mapping(M_PL_011_graph)$type
# convert network to incidence matrix
M_PL_011_matrix <- get.incidence(M_PL_011_graph)5.2.2 The nestedness function
Now that we have downloaded our network, we measure its nestedness. We will be using the same function you used in previous sessions:
compute_nestedness <- function(B){
# Get number of rows and columns
nrows <- nrow(B)
ncols <- ncol(B)
# Compute nestedness of rows
nestedness_rows <- 0
for(i in 1:(nrows-1)){
for(j in (i+1): nrows){
c_ij <- sum(B[i,] * B[j,]) # Number of interactions shared by i and j
k_i <- sum(B[i,]) # Degree of node i
k_j <- sum(B[j,]) # Degree of node j
if (k_i == 0 || k_j==0) {next} # Handle case if a node is disconnected
o_ij <- c_ij / min(k_i, k_j) # Overlap between i and j
nestedness_rows <- nestedness_rows + o_ij
}
}
# Compute nestedness of columns
nestedness_cols <- 0
for(i in 1: (ncols-1)){
for(j in (i+1): ncols){
c_ij <- sum(B[,i] * B[,j]) # Number of interactions shared by i and j
k_i <- sum(B[,i]) # Degree of node i
k_j <- sum(B[,j]) # Degree of node j
if (k_i == 0 || k_j==0) {next} # Handle case if a node is disconnected.
o_ij <- c_ij / min(k_i, k_j) # Overlap between i and j
nestedness_cols <- nestedness_cols + o_ij
}
}
# Compute nestedness of the network
nestedness <- (nestedness_rows + nestedness_cols) / ((nrows * (nrows - 1) / 2) + (ncols * (ncols - 1) / 2))
return(nestedness)
}5.2.3 Computing nestedness
Let’s compute the nestedness value for the network we downloaded before, as in previous sessions. Next, we will implement the null models.
# Calculate the nestedness value for our network and store it
nestedness_M_PL_011 <- compute_nestedness(M_PL_011_matrix)
nestedness_M_PL_011## [1] 0.6020715.2.4 Equifrequent null model
First, we’ll start with the equifrequent null model. This is the function we will use to run the model. It takes as arguments, the matrix you wish to use (\(d\)) and the number of replicates (\(t_{max}\)). The function performs \(t_{max}\) randomizations of \(d\) and computes the associated nestedness value of each randomisation. The function outputs a vector of length \(t_{max}\) with our nestedness estimates.
equifrequent_model <- function(d,t_max){
rows <- nrow(d)
columns <- ncol(d)
t <- 1
null_nest = rep(NA,t_max)
while (t < t_max + 1) {
B <- matrix(0, rows, columns)
number_ones <- nnzero(d)
count_ones <- 0
while (count_ones < number_ones) {
x <- ceiling(runif(1, min = 0, max = rows))
y <- ceiling(runif(1, min = 0, max = columns))
while (B[x,y] == 1) {
x <- ceiling(runif(1, min = 0, max = rows))
y <- ceiling(runif(1, min = 0, max = columns))
}
B[x,y] <- 1
count_ones <- count_ones + 1;
}
# Remove unconected species if present
B = trim_network(B)
null_nest[t] = compute_nestedness(B)
t<-t+1
}
# unlist results
null_nest <- unlist(null_nest)
return(null_nest)
}
trim_network = function(network){
# Removes any columns or rows without interactions
network <- network[,colSums(network != 0) > 0]
network <- network[rowSums(network !=0) > 0,]
return(network)
}Let’s run the equifrequent null model with 100 replicates on our downloaded network. Run the command below in your Rscript file, make sure to add the required parameter values.
# Run the equifrequent model for our network with 100 replicates
nestedness_equifrequent_list <- equifrequent_model(d = , t_max = )Now that we have our nestedness estimates, we need to estimate the significance of the nested value we initially observed using the z-score. The formula to estimate z-scores is:
\[ z-score = \frac{observed\ nestedness - mean(nestedness)}{sd(nestedness)} \]
To calculate the z-score, we first have to obtain the mean and the standard deviation of the nestedness values we obtain from our null model. The mean and standard deviation are calculated in R with the mean and sd functions, respectively. Run the command below in your Rscript file, make sure to add the required parameter values.
# Compute mean for nestedness values estimated by null model
mean_nestedness <- mean()
# Compute sd for nestedness values estimated by null model
sd_nestedness <- sd()Now we can calculate the z-score for the null model. Run the command below in your Rscript file, make sure to add the required parameter values.
# Compute z score following formula
z_score <- Finally we can calculate the probability associated to the z-score using the pnorm and setting the lower.tail parameter to FALSE. Run the command below in your Rscript file, make sure to add the required parameter values.
# Compute associated p value
p_val <-pnorm(, lower.tail = FALSE)
# Print p value
print(p_val)5.2.5 Probabilistic cell model
Now let’s move to the probabilistic cell model. This is the function we will use to implement it. As before, it takes as arguments, the matrix you wish to use (\(d\)) and the number of replicates (\(t_{max}\)). The function performs \(t_{max}\) randomizations of \(d\) and computes the associated nestedness value of each randomisation. The function outputs a vector of length \(t_{max}\) with our nestedness estimates.
cell_model <- function(d,t_max){
rows <- nrow(d)
columns <- ncol(d)
t<-1
null_nest = rep(NA,t_max)
while (t <= t_max + 1){
PR <- matrix(0, rows, 1)
PC <- matrix(0, columns, 1)
B <- matrix(0, rows, columns)
for (i in 1:rows){
number_ones<-0
for (j in 1:columns){
if( d[i,j] == 1){
number_ones<-number_ones+1
}
}
PR[i] <- number_ones/columns
}
for (j in 1:columns){
number_ones<-0
for (i in 1:rows){
if( d[i,j] == 1){
number_ones<-number_ones+1
}
}
PC[j] <- number_ones/rows
}
for (i in 1:rows){
for (j in 1:columns){
p <- ( PR[i]+PC[j] )/2;
r <- runif(1)
if( r < p ){
B[i,j] <- 1;
}
}
}
B = trim_network(B)
null_nest[t] = compute_nestedness(B)
t<-t+1
}
# unlist results
null_nest <- unlist(null_nest)
return(null_nest)
}Repeat the same process we followed for the equifrequent null model, but now using the probabilistic cell model. You should do the following:
- Run the null model 100 times with using the
M_PL_011_matrix. - Compute the mean and standard deviation of our null model estimates.
- Calculate the z-score.
- Obtain an associated p-value.
# Run the probabilistic cell model in your own Rscript5.2.6 Swap model
Lastly, we focus on the swap model. This is the function we will use to compute the null model. As before, it takes as arguments, the matrix you wish to use (\(d\)) and the number of replicates (\(t_{max}\)). The function performs \(t_{max}\) randomizations of \(d\) and computes the associated nestedness value of each randomisation. The function outputs a vector of length \(t_{max}\) with our nestedness estimates.
swap_model = function(d,t_max){
# Make sure we are working with a matrix
d = as.matrix(d)
rows <- nrow(d)
columns <- ncol(d)
swap <- 0;
steps <- t_max;
swap_nest = rep(NA,t_max)
while (swap < steps) {
i <- ceiling(runif(1, min = 0, max = rows))
j <- ceiling(runif(1, min = 0, max = columns))
while (d[i,j] == 0) {
# for the first pair of numbers
i <- ceiling(runif(1, min = 0, max = rows))
j <- ceiling(runif(1, min = 0, max = columns))
}
# for the second pair of numbers
q <- ceiling(runif(1, min = 0, max = rows))
z <- ceiling(runif(1, min = 0, max = columns))
# different row and column
while ((q == i) || (q == j)){
q <- ceiling(runif(1, min = 0, max = rows))
}
# different row and column
while ((z == i) || (z ==j)){
z <- ceiling(runif(1, min = 0, max = columns))
}
while (d[q,z] == 0) {
q <- ceiling(runif(1, min = 0, max = rows))
while ((q == 1) || (q == j)) {
q <- ceiling(runif(1, min = 0, max = rows))
}
z <- ceiling(runif(1, min = 0, max = columns))
while ((z == i) || (z ==j)){
z <- ceiling(runif(1, min = 0, max = columns))
}
}
if (d[i,z] == 0 && d[q,j] == 0) {
d[i,j] <- 0;
d[q,z] <- 0;
d[i,z] <- 1;
d[q,j] <- 1;
d = trim_network(d)
swap_nest[swap+1] = compute_nestedness(d)
swap <- swap + 1;
}
}
# unlist results
swap_nest <- unlist(swap_nest)
return(swap_nest)
}Repeat the same process we followed for the other two null models, but now using the swap model. You should do the following:
- Run the null model 100 times with using the
M_PL_011_matrix. - Compute the mean and standard deviation of our null model estimates.
- Calculate the z-score.
- Obtain an associated p-value.
# Run the swap model in your own Rscript5.3 Computer part II
In this final section, we will use the null models to test whether a set of pollination networks are significantly more or less nested than a set of seed dispersal networks. We will use null models to answer this question.
5.3.1 Code
To save some time, I have already downloaded the necessary networks. The first thing we need to is to load the networks. The seed dispersal networks are stored as entries in the list called seed_networks which is found in the seed_networks.Rdata file. The pollination networks are stored as entries in the list called pollination_networks which is found in the pollination_networks.Rdata file. We will begin by loading these two files.
# Load pollination networks
load("~/ecological_networks_2022/downloads/Data/03-24_null_models/pollination_networks.Rdata")
# Load seed networks
load("~/ecological_networks_2022/downloads/Data/03-24_null_models/seed_networks.Rdata")Now that we have our networks, we can continue to run the null models, in this example we will use the cell null model. Let’s outline our worflow:
- First, compute the nestedness value of each of the networks in the
seed_networksandpollination_networkslists. - Next, run the cell null model with 20 replicates for each network in each type of network.
- Finally, run a t-test to determine whether there are differences between the two types of networks in relation to their standardized nestedness values.
Below you will find the code to perform the workflow outlined above. However, if you’re up for a challenge, feel free to write your own code. Use the p-value you obtain to answer the question at the bottom of this exercise.
#################################################
# Compute z-score for seed-disperser networks
# Create vector to store z-scores
z_score_vector_seed_nets <- rep(0,length(seed_networks))
# Loop through the seed-disperser networks
for (network in 1:length(seed_networks)) {
# Set the current network
current_seed_net <- as.matrix(seed_networks[[network]])
# Compute nestedness for current network
current_nestedness <- as.numeric(compute_nestedness(current_seed_net))
# Run cell null model with current network and 20 replicates
z_score_vector <- cell_model(current_seed_net,20)
# Compute mean of zscores
mean_z_score <- mean(z_score_vector)
# Compute standard devation of zscores
sd_z_score <- sd(z_score_vector)
# Compute the zscore for the network
z_score_seed_nets <- (current_nestedness - mean_z_score)/sd_z_score
# Store zscore in vector
z_score_vector_seed_nets[network] <- z_score_seed_nets
}
#################################################
# Compute z-score for pollination networks
# Create vector to store z-scores
z_score_vector_pollination_nets <- rep(0,length(pollination_networks))
# Loop through the seed-disperser networks
for (network in 1:length(pollination_networks)) {
# Set the current network
current_seed_net <- as.matrix(seed_networks[[network]])
# Compute nestedness for current network
current_nestedness <- as.numeric(compute_nestedness(current_seed_net))
# Run cell null model with current network and 20 replicates
z_score_vector <- cell_model(current_seed_net,20)
# Compute mean of zscores
mean_z_score <- mean(z_score_vector)
# Compute standard devation of zscores
sd_z_score <- sd(z_score_vector)
# Compute the zscore for the network
z_score_pollination_nets <- (current_nestedness - mean_z_score)/sd_z_score
# Store zscore in vector
z_score_vector_pollination_nets[network] <- z_score_pollination_nets
}
# Calculate t-test to compare vector of zscores
t.test(z_score_vector_seed_nets,z_score_vector_pollination_nets)